ABOVE: © SCIENCE PHOTO LIBRARY, KEITH CHAMBERS
When microbiologist Breck Duerkop started his postdoc in 2009, he figured he’d be focusing on bacteria. After all, he’d joined the lab of microbiome researcher Lora Hooper at the University of Texas Southwestern Medical Center in Dallas to study host-pathogen interactions in the mammalian gut and was particularly interested in what causes some strains of normally harmless commensal bacteria, such as Enterococcus faecalis, to become dangerous, gut-dominating pathogens. He’d decided to explore the issue by giving germ-free mice a multidrug-resistant strain of E. faecalis that sometimes causes life-threatening infections in hospital patients, and analyzing how these bacteria express their genes in the mouse intestine.
Not long into the project, Duerkop noticed something else going on: some of the genes being expressed in E. faecalis weren’t from the regular bacterial genome. Rather, they were from bacteriophages, bacteria-infecting viruses that, if they don’t immediately hijack and kill the cells they infect, can sometimes incorporate their genetic material into the bacterial chromosome. These stowaway viruses, known as prophages while they’re in the bacterial chromosome, may lie dormant for multiple bacterial generations, until certain environmental or other factors trigger their reactivation, at which point they begin replicating and behaving like infectious agents once again. (See illustration below.) Duerkop’s data showed that the chromosome of the E. faecalis strain he was using contained seven of these prophages and that the bacteria were churning out virus particles with custom combinations of these prophage sequences during colonization of the mouse gut.
The presence of viruses in Duerkop’s E. faecalis strain wasn’t all that surprising. Natural predators of bacteria, bacteriophages are the most abundant biological entities on the planet, and in many fields, researchers take their presence for granted. “Nobody really was thinking about phages in the context of bacterial communities” in animal hosts, Duerkop says. “It would [have been] very easy to just look at it and say, ‘Oh, there are some phage genes here. . . . Moving on.’” But he was curious about why E. faecalis would be copying and releasing them, rather than leaving the prophages asleep in its chromosome, while it was trying to establish itself in the mouse intestine.
Predation is just one type of phage-bacteria interaction taking place within the mammalian microbiome; many phages are capable of inserting their genomes into the bacterial chromosome.
Encouraged by Hooper, he put his original project on hold in order to dig deeper. To his surprise, he discovered that the E. faecalis strain, known as V583, seemed to be using its phages to gain a competitive advantage over related strains. Experiments with multiple E. faecalis strains in cell culture and in mice showed that the phage particles produced by the bacteria didn’t harm other V583 cells, but infected and killed competing strains. Duerkop and his colleagues realized that, far from being background actors in the bacterial community, the phages “are important for colonization behavior” for this opportunistic pathogen.
The idea that a phage could play such a significant role in the development of the gut bacterial community was relatively novel when the team published its results in 2012. Since then, “it’s been pretty well established that phages can shape the assembly of microbial communities in the intestine, and that can influence the outcome on the host—either beneficially or detrimentally,” says Duerkop, who now runs his own lab at the University of Colorado School of Medicine in Aurora. There’s evidence that phages help bacteria share genetic material with one another, too, and may even interact directly with the mammalian immune system, an idea that Duerkop says would have had you “laughed out of a room” of immunologists just a few years ago.
Predation by phages influences bacterial communities
Around the time that Duerkop was working on E. faecalis in Dallas, University of Oxford postdoc Pauline Scanlan was studying Pseudomonas fluorescens, a bacterial species that is abundant in the natural environment and is generally harmless to humans, although it’s in the same genus as the important human pathogen Pseudomonas aeruginosa. Bacteria in this genus sometimes evolve what’s known as a mucoid phenotype—that is, cells secrete large amounts of a compound called alginate, forming a protective goo around themselves. In P. aeruginosa, this goo can help the bacteria evade the mammalian immune system and antibiotics, and “when it crops up, it’s not good news” for the patient, Scanlan says. She was curious about what causes a non-mucoid bacterial population to evolve into a mucoid one and had found previous research suggesting that the presence of bacteriophages could play a role. Other studies documented high densities of phages in mucus samples from the lungs of some cystic fibrosis patients with P. aeruginosa infections.
Working in the lab of evolutionary biologist Angus Buckling (now at the University of Exeter), Scanlan grew a strain of P. fluorescens with a phage called Phi2 that specifically infects and destroys this bacterium. Cells with the gummy mucoid coating, the researchers noted, were more resistant to phage infection than regular cells were. What’s more, over generations, bacterial populations were more likely to evolve the mucoid phenotypes in the presence of Phi2 than they were in its absence, indicating that the phenotype may arise in Pseudomonas as an adaptive response to phage attack. Scanlan, now at University College Cork (UCC) in Ireland, notes that more work is needed to extend the findings to a clinical setting, but the results hint that phages could in some cases be responsible for driving bacteria to adopt more virulent phenotypes.
Such a role for viruses in driving bacterial evolution fits well with phages’ reputation as “the ultimate predators,” says Colin Hill, a molecular microbiologist also at UCC who got his introduction to phages studying bacteria used in making fermented foods such as cheese. Hill notes an estimate commonly cited in the context of marine biology—a field that explored phage-bacteria interactions long before human biology did—that phages kill up to 50 percent of the bacteria in any environment every 48 hours. “The thing that any bacterium has on its mind most, if bacteria had minds, would be phage,” Hill says, “because it’s the thing most likely to kill them.”
Several in vivo animal studies lend support to the idea that predatory phages help shape bacterial evolution and community composition in the mammalian microbiome. In 2019, for example, researchers at Harvard Medical School reported that phages not only directly affect the bacteria they infect in the mouse gut, but also influence the rest of the microbiome community via cascading effects on the chemical and biological composition of the gut. Observational studies hint at similar processes at work in the human gut. A few years ago, researchers at Washington University Medical School in St. Louis observed patterns of phage and bacterial population dynamics that resembled predator-prey cycles in the guts of children younger than two years old: low bacterial densities at birth were followed by decreases in phages, after which the bacteria would rebound, and then the phages would follow suit. The team concluded that these cycles were likely a natural part of healthy microbiome development.
Although researchers are only just beginning to appreciate the importance of phages in microbiome dynamics, they’ve already begun to explore links to human disease. Authors of one 2015 study reported that Crohn’s disease and ulcerative colitis patients showed elevated levels of certain phages, particularly within the viral order Caudovirales. They proposed that an altered virome could contribute to pathogenesis through predator-prey interactions between phages and their bacterial hosts. Other studies have explored possible phage-driven changes in the bacterial community in human diseases such as diabetes and certain cancers that are known to be associated with a disrupted microbiome. But the observational nature of human microbiome studies prevents conclusions about what drives what—changes in virome composition could themselves be the result of disruptions to the bacterial community, for example.
Currently, researchers are exploring the possibility of using predatory phages as weapons against pathogenic bacteria, particularly those that present a serious threat to public health due to the evolution of resistance to multiple antibiotics. It’s the principle that “the enemy of my enemy is my friend,” says Yale University virologist and evolutionary biologist Paul Turner. “If we have a pathogen that is in your microbiome, can we go in and remove that bacterial pathogen by introducing a predatory phage, something that is cued to only destroy [that pathogen]?” Although the strategy was first proposed more than a century ago, “we and others are trying to update it,” he adds. (See “My Enemy’s Enemy” below.)
PHAGE LIFECYCLEPhages can interact with bacteria in two main ways. In the first, phages infect a bacterial cell and hijack that cell’s protein-making machinery to replicate themselves, after which the newly made virus particles lyse the bacterium and go on to infect more cells. In the second process, known as lysogeny, the viral genome is incorporated into the bacterial chromosome, becoming what’s known as a prophage, and lies dormant—potentially for many generations—until certain biotic or abiotic factors in the bacterium or the environment induce it to excise itself from the chromosome and resume the cycle of viral replication, lysis, and infection of new cells.  © LISA CLARK |
Phages provide a gene-delivery service for bacteria
Predation is just one type of phage-bacteria interaction taking place within the mammalian microbiome. Many phages are capable of inserting their genomes into the bacterial chromosome, a trick beyond the bounds of traditional predator-prey relationships in other kingdoms of life that adds complexity to the relationship between phages and bacteria, and consequently, to phages’ potential influences on human health.
This role for phages has long been of interest to Imperial College London’s José Penadés. Over the last 15 years or so, he and colleagues have described various ways in which many phages help bacteria swap genetic material among cells. He likens phages to cars that bacteria use to transport cargo around and says that, in his opinion, it almost makes sense to view phages as an extension of bacteria rather than as independent entities. “This is part of the bacterium,” he says. “Without phages, bacteria cannot really evolve. They are absolutely required.”
“With lateral [transduction] you can move huge parts of the bacterial chromosome.”
—José Penadés, Imperial College London
In the simplest case, the genetic material being transported consists of viral genes in the genomes of so-called temperate phages, which spend at least part of their lifecycle stashed away in bacterial chromosomes as prophages. These phages are coming to be appreciated by microbiologists as an important driver of bacterial evolution in the human microbiome, notes Hill. The lack of practical and accurate virus detection methods makes it difficult to precisely characterize a lot of the phages resident in mammalian guts, but microbiologists estimate that up to 50 percent are temperate phages, and, more importantly for human health, that many of them may carry genes relevant to bacterial virulence. Researchers have long known, for example, that many toxins produced by bacteria—including Shiga toxin, made by some pathogenic E. coli strains, and cholera toxin, secreted by the cholera-causing bacterium Vibrio cholerae—are in fact encoded by viral genes contained in the bacterial chromosome, and that infection by temperate phages that carry these genes may be able to turn a harmless bacterial population into one that’s pathogenic.
Evidence from other studies points to phages as capable of transporting not just their own genomes, but bits of bacterial DNA as well. In the best-studied examples of this phenomenon, known as bacterial transduction, tiny chunks of the bacterial genome get packed up into viral particles instead of or alongside the phage genome, and are shuttled to other bacterial cells. In 2018, however, Penadés and colleagues presented results showing that very large pieces of bacterial DNA can also be exchanged this way, in a process the team named lateral transduction. Not only does the discovery have implications for how researchers understand viral replication in infected cells, it shines light on a novel way for bacteria to share their genes. “With lateral [transduction] you can move huge parts of the bacterial chromosome,” says Penadés. The team first observed the phenomenon in the important human pathogen Staphylococcus aureus, and is now looking for it in other taxa, he adds. “Right now, for us, it’s important to show that it’s a general mechanism, with many bugs involved.”
Although the research is still in the nascent stages, this mechanism could help explain findings from University of Barcelona microbiologist Maite Muniesa and others who have been studying whether phages transport antibiotic resistance genes between bacterial cells, and whether they can act as reservoirs for these genes in the natural environment. Early studies on this issue had proposed that, like many toxin genes, antibiotic resistance genes might be encoded in viral sequences and thus transported to bacteria with the rest of the viral genome. But the idea wasn’t without controversy—a 2016 analysis of more than 1,100 phage genomes from various environments concluded that phage genomes only rarely include antibiotic resistance genes. That study’s authors argued that prior reports of these genes in phage genomes were likely due to contamination, or to the difficulty of distinguishing viral sequences from bacterial ones.
Nevertheless, Muniesa’s team has published multiple reports of antibiotic resistance sequences in phage particles, including in samples of meat products from a Barcelonan fresh-food retailer, and more recently in seawater samples—not only from the Mediterranean coastline but even off the coast of Antarctica, far from human populations that use antibiotics. “We were pretty surprised that we found these particles in this area with low human influence,” Muniesa says. Although her team hasn’t determined whether the antibiotic resistance sequences are of phage or bacterial origin, she suspects they might be bacterial genes that ended up in phage particles during lateral transduction or some process like it. “Bacteria are using these phage particles in a natural way to move [genes] between their brothers and sisters, let’s say,” she says. “It’s happening everywhere.”
Duerkop cautions that it’s not yet clear how often phage-mediated transfer of antibiotic resistance genes occurs or how significant it is in the epidemiology of drug-resistant infections in people. “It’s not to say that antibiotic resistance can’t be mediated through phage,” he says. “I just don’t think it’s a major driver of antibiotic resistance.”
Whatever its natural role, temperate phages’ ability to insert themselves into bacterial genomes could have applications in new antibacterial therapies. Viruses that insert pathogenicity-reducing genes or disrupt the normal expression of the bacterial chromosome could be used to hobble dangerous bacteria, for example—an approach that proved successful last year in mouse experiments with Bordetella bronchiseptica, a bacterium that often causes respiratory diseases in livestock. Using a phage from the order Siphoviridae, researchers found that infected B. bronchiseptica cells were substantially less virulent in mice than control cells were, likely because the viral genome had inserted itself in the middle of a gene that the bacterium needs to infect its host. What’s more, injecting mice with the phage before exposing them to B. bronchiseptica seemed to completely protect them from infection by the microbe, hinting at the possibility of using temperate phages as vaccines against some bacteria.
GUT WARSBacteria-infecting viruses, or bacteriophages, may influence microbial communities in the mammalian gut in various ways, some of which are illustrated here. Through predation, phages can influence the abundance of specific bacterial taxa, with indirect effects on the rest of the community, and can drive the evolution of specific bacterial phenotypes. Phages can also incorporate their genomes into bacterial chromosomes, where the viral sequences lie dormant as prophages until reactivated. Researchers have found that phages interact directly with mammalian cells in the gut, too. These cross-kingdom interactions could affect the health of their eukaryotic hosts. 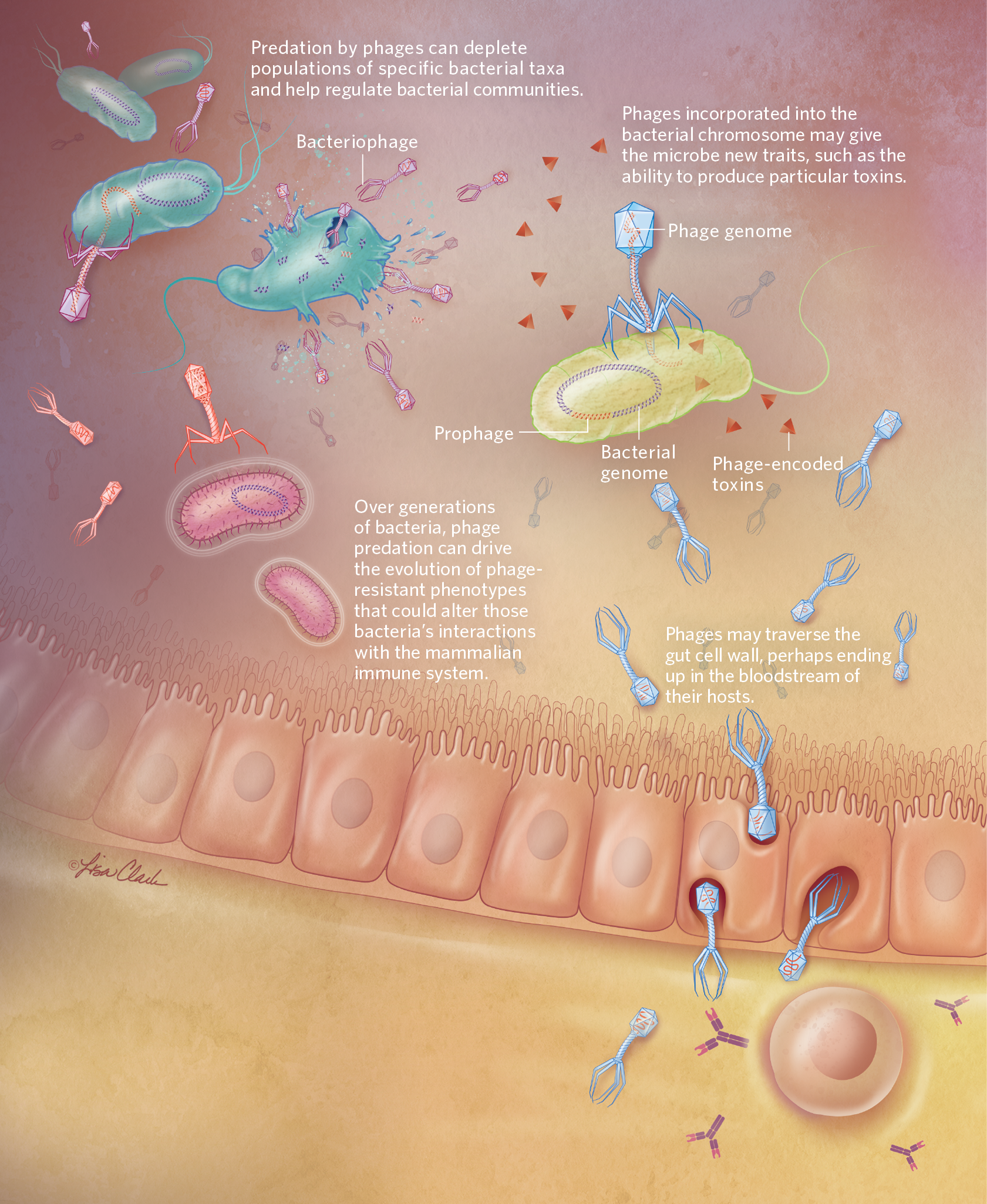 © LISA CLARK 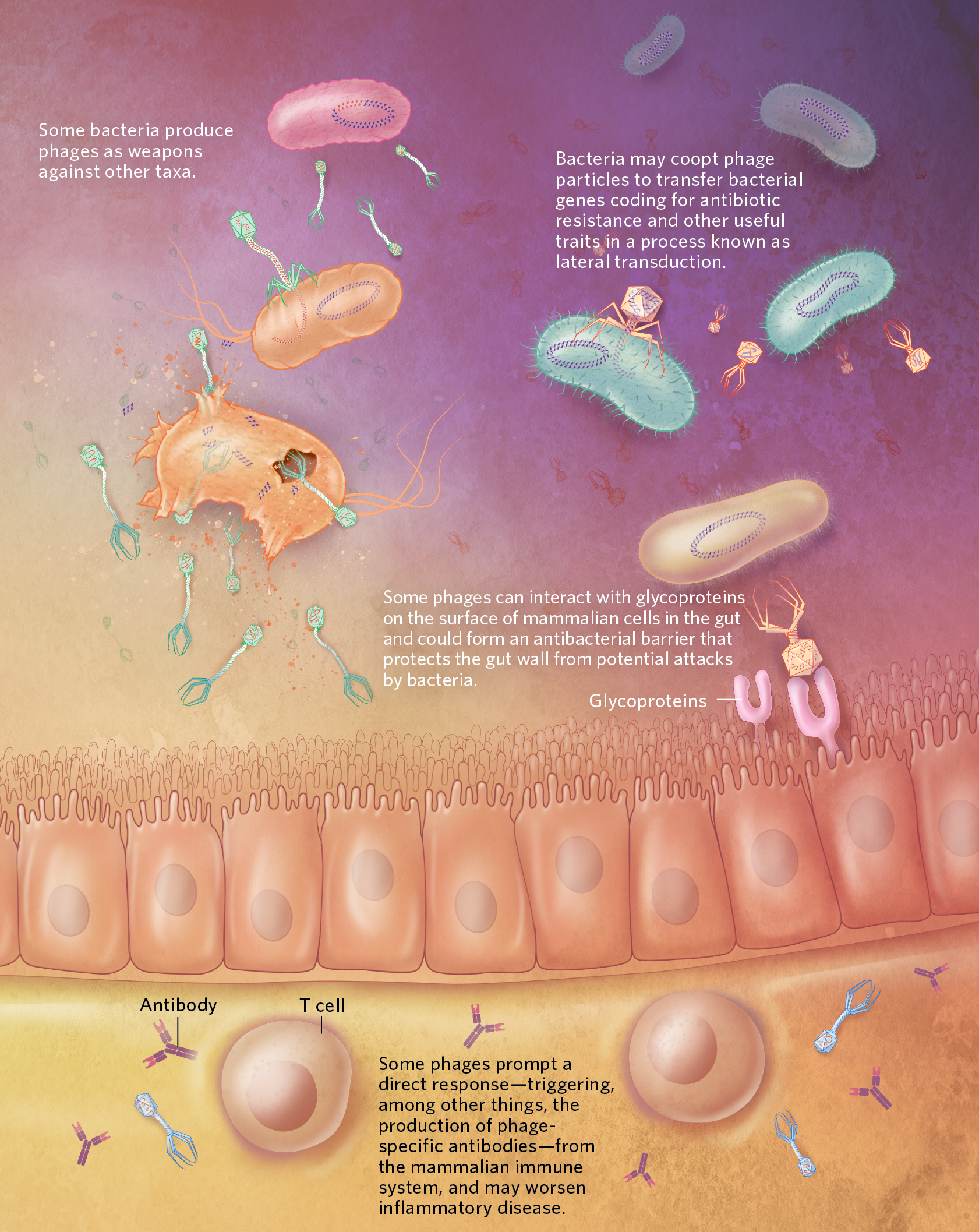 © LISA CLARK |
Direct interactions between bacteriophages and mammalian cells
Despite growing interest in phages’ role in shuttling material among bacteria, some of the biggest recent developments in research on phages in the human gut have turned out not to involve bacteria at all. One of the key pieces of this particular puzzle was fitted by University of Utah microbiologist June Round and her colleagues, who as part of a phage therapy study a few years ago fed several types of Caudovirales phages to mice that were genetically predisposed to certain types of cancer and had been infected with a strain of E. coli known to increase that risk. “The premise was pretty simplistic,” recalls Round. “It was just to identify a cocktail of phage that would target bacteria that we know drive chronic colorectal cancer.”
The team was surprised to see that the phages, despite being viewed by most researchers as exclusively bacteria-attacking entities, prompted a substantial response from the mice’s immune systems—mammalian defenses that should, according to conventional wisdom, be indifferent to the war between bacteria and phages in the gut. Intrigued, the researchers tried adding their phage cocktail to mice that had had their gut bacteria completely wiped out with antibiotics. Still, they saw an immune response. It was then, Round says, that “we realized that [the phages] were likely interacting with the immune system.”
Exploring further, the team found that the phages were activating both innate and adaptive immune responses in mice. In rodents with colitis, the phages exacerbated inflammation. Turning their attention to people, the researchers isolated phages from ulcerative colitis patients with active disease, as well as from patients with disease in remission and from healthy controls, and showed that only viruses collected from patients with active disease stimulated immune cells in vitro. And when the team studied patients who received fecal microbiota transplantation—an experimental treatment for ulcerative colitis that involves giving beneficial gut bacteria to a patient to try to alleviate inflammation and improve symptoms—the researchers found that a lower abundance of Caudovirales in a recipient’s intestine at the time of transplant correlated with treatment success.
Some of the biggest recent developments in research on phages in the human gut have turned out not to involve bacteria at all.
By the time the team published its results in 2019, a couple of other groups had also documented evidence of direct interactions between phages and host immune systems. Meanwhile, Duerkop, Hooper, and colleagues reported that mice with colitis tended to have specific bacteriophage communities, rich in Caudovirales, that developed in parallel with the disease. Many of the types of phage they identified in the intestines of those diseased mice also turned up in high abundance in samples taken from the guts of people with inflammatory bowel disease, the researchers noted in their paper, supporting a possible role for phages in the development of disease.
Round says that researchers are still unsure about exactly why these trans-kingdom interactions are happening—particularly when it comes to host adaptive immune responses, which tend to be specific to a particular pathogen. She speculates that mammalian hosts might derive a benefit from destroying certain phages if those phages are carrying genes that could aid a bacterium with the potential to cause disease. Exactly how immune cells would detect what genes a phage is carrying isn’t yet clear.
Meanwhile, hints of collaboration between eukaryotic cells and phages have cropped up in the work of several other labs. One recent study of a phage therapy against P. aeruginosa found that phages and immune cells seem to act in synergy to clear infections in mice. Other work has indicated that phages bind to glycoproteins presented by cells along the mucosal surfaces of the mammalian gut and may provide a protective barrier against bacterial pathogens—a relationship that some microbiologists have argued represents an example of phage-animal symbiosis. Duerkop adds that there’s evidence emerging to support the idea that phages in the mammalian intestine not only can be engulfed by certain eukaryotic cells, but also might slip out of the gut and into the bloodstream to make their way to other parts of the body, with as yet undiscovered consequences.
Whether these mechanisms can be exploited for therapeutic purposes remains to be seen, but Round notes that they do raise the possibility of unintended effects in some circumstances if researchers try to use phages to influence human health via the gut microbiome. At least in the type of chronic inflammatory diseases she and her team have been studying, “we might just be making it worse” by using phages to target disease-causing bacteria, she says, adding that all research groups studying such approaches should take into account potential knock-on effects. Considering phages’ multiple interactions, with both bacteria and animal cells, she says, “it’s a lot more complex than what we’d appreciated.”
My Enemy’s EnemyBacteriophages’ ability to selectively target and kill specific bacterial strains has long been recognized as a possible basis for antimicrobial therapies. Proposed by researchers in Europe as early as 1919, phage therapy went on to be widely promoted in Germany, the USSR, and elsewhere before being overtaken worldwide by the soaring popularity of antibiotics in the 1940s. But the strategy has come back into fashion among many microbiologists, thanks to the growing public health problem of antibiotic resistance in bacterial pathogens and to the rapidly improving scientific understanding of phage-bacteria interactions. See “Bacteriophage Boom?”Some of the latest approaches aim not only to target specific bacteria with phages, but also to avoid (or exploit) the seemingly inevitable evolution of phage resistance in those bacteria. One way researchers try to do this is by taking advantage of an evolutionary trade-off: bacterial strains that evolve adaptations to one therapy will often suffer reduced fitness when confronted with a second therapy, perhaps one that targets the same or similar pathways in a different way.  © ISTOCK.COM, PETERSCHREIBER.MEDIA Yale University virologist and evolutionary biologist Paul Turner, for example, has studied how phages in the Myoviridae (a family in the order Caudovirales) can promote antibiotic sensitivity in the important human pathogen Pseudomonas aeruginosa. Turner and colleagues showed a few years ago that one such phage binds to a protein called OprM in the bacterial cell membrane, and that bacterial populations under attack from these phages will often evolve reduced production of OprM proteins as a way of avoiding infection. However, OprM also happens to be important for pumping antibiotics out of the cell, such that abnormal OprM levels can reduce bacteria’s ability to survive antibiotic treatment in vitro. A handful of groups have published case studies using this kind of approach, known as phage steering, in humans. A couple years ago, for example, Turner and colleagues reported that a post-surgery patient’s chronic P. aeruginosa infection cleared up after treatment with the OprM-binding phage and the antibiotic ceftazidime. Researchers at the University of California, San Diego, in partnership with California-based biotech AmpliPhi Biosciences (now Armata Pharmaceuticals), reported similar success in a cystic fibrosis patient with a P. aeruginosa infection who was treated with a mixture of phages and with antibiotics. A Phase 1/2 trial for that therapy was greenlighted by the US Food and Drug Administration last October. The complexity of the relationship between phages and bacteria, not to mention recently discovered interactions between phages and eukaryotic cells, has many researchers tempering optimism about phage therapy with caution. “There might be off-target effects to this that we hadn’t really thought about,” says University of Colorado School of Medicine microbiologist Breck Duerkop. That said, thanks to research in the last few years, “the black veil on phage therapy is, I believe, being lifted,” he adds, “which I’m really excited about because I think they have a ton of potential to be used in biomedicine.” |








